For a brief overview of what a package is, click here.
Note that if you've obtained an existing package and you're only going to be building models that use those existing cell types, you might want to start with the next section, which focuses on building models. (It does refer back to this section for some concepts.) At some point you will probably want to make modifications to the cell types, at which point you will want to come back and study this section.
- Developing Packages of Cell Types
- Files in a Package Directory
- Defining Properties and Fields
- Writing Kernels
Files in a Package Directory
A package consists of a set of MATLAB ".m" files and C++ ".h" files which together define a set of cell types. These files must all reside in a single directory (the "package directory"). See here for an overview.
Now would be a good time to have a look at the files in cns/demopkg, or one of the other available packages.
The following table lists the source files that make up a package and the kind of information each contains.
| File Name | Purpose / Contents | Format |
| package.m |
|
Details. |
| package.h |
|
Free-format C++ code. |
| package_base.m |
|
Details. |
| package_base.h | Free-format C++ code. | |
| package_type.m |
|
Details. |
| package_type.h | Free-format C++ code. |
Packages must be compiled with the cns_build command. Compiling a package produces several files starting with "package_compiled_".
Note: whenever you change any of the source files in a package directory, you must re-run the cns_build command.
A package directory can contain other files; CNS ignores any file not starting with "package_".
Defining Properties and Fields
".m" File Format
Everything about a package -- its cell types, properties, fields, etc. -- except the compute kernels -- is defined in the package's ".m" files.
The main package.m file looks like this:
classdef package < cns_package methods (Static) %------------------------------------------------------------------ function f = CNSFields % Define any model-wide fields here. (Details.) % Define any model-wide compile-time constants here. (Details.) end %------------------------------------------------------------------ % Any package-level MATLAB methods you may wish to add. (Details.) %------------------------------------------------------------------ end end |
Note that while we use MATLAB's classdef syntax, we do not make much use of MATLAB's object-oriented programming model. It is simply a convenient syntax for defining a type hierarchy. All the methods are static. We do not create MATLAB objects to represent models, layers, or cells. Rather, we build a model structure which CNS uses to instantiate a model on the GPU. Models, and the layers and cells that comprise them, live in GPU memory, and we use cns commands to access them from within MATLAB.
The ".m" file for the base type (package_base.m) must start like this:
classdef package_base < cns_base ... |
Otherwise it is the same as the ".m" file for any other, more specific cell types, which are defined as follows. Note that most of a cell type's definitions will be inherited by any subtypes of that type.
classdef package_type < package_supertype methods (Static) %------------------------------------------------------------------ function p = CNSProps % Define any type-specific properties here. (Details.) end %------------------------------------------------------------------ function f = CNSFields % Define any type-specific fields here. (Details.) % Define any type-specific compile-time constants here. (Details.) end %------------------------------------------------------------------ % Any type-specific MATLAB methods you may wish to add. (Details.) %------------------------------------------------------------------ end end |
When creating a new cell type, it's often most convenient to copy the ".m" file of another type and modify it.
Type Properties
The following table lists the properties you can define for a cell type.
Each property is defined by adding a line like this to the CNSProps method of the type's ".m" file.
p.propname = value;
For example:
p.abstract = true;
| Property Name | Usage |
| abstract | Defaults to false. If set to true, this means the type exists only to be a supertype of other types. In a CNS model, you cannot create a layer of cells of an abstract type, only a subtype.
The "base" type of a package is often abstract. |
| dnames dims dparts dmap |
Together, these properties define the dimensionality of a layer of cells of this type. Click here for details. |
| synType | If cells of this type are going to explicitly enumerate their input cells, use this property to specify which type of cells those inputs will be. This tells CNS what fields will be available to read from the input cells. You will want to be as specific as possible. If any type of cell can be an input, just specify p.syntype = 'base'. However, this would mean you could only read fields defined for all cell types, i.e. defined at the 'base' level. |
Defining Dimensionality
All non-abstract cell types must define their dimensionality, i.e.:
- how many dimensions will a layer of such cells have?
- what are the names of those dimensions?
- how should those dimensions be represented using the GPU's two internal dimensions?
- should cell positions within a layer be mapped to common coordinates?
Consider the following example:
p.dnames = {'f' 't' 'y' 'x'};
p.dims = {1 2 1 2};
p.dparts = {2 2 1 1};
Layers of this cell type will be 4-D; it will take four coordinates to identify a particular cell. In CNS you will be able to refer to the dimensions by their names ('f', 't', 'y', 'x'). dims and dparts tell CNS how each of the four dimensions is represented in the GPU's two dimensions. Here, dimensions 'f' and 'y' are assigned to internal dimension 1, with 'f' as the "outer" dimension (part 2) and 'y' as the "inner" dimension (part 1). This means that cells with adjacent 'y' values will be stored next to each other in GPU memory, while cells with adjacent 'f' values may be far apart. Dimensions 't' and 'x' are similarly assigned to internal dimension 2.
The choice of inner and outer dimensions has performance consequences: if some other cell is pooling over cells in this layer using nested for loops, the inner loops should correspond to the inner dimensions.
Here is an example of the simpler case where there are only two dimensions.
p.dnames = {'y' 'x'};
p.dims = {1 2};
p.dparts = {1 1};
You can even have just one dimension; for example:
p.dnames = {'f'};
p.dims = {1};
p.dparts = {1};
Performance note: CNS becomes inefficient when the size of internal dimension 1, for a layer, is small relative to the first element of #BLOCKSIZE. For example, if your layer is represented internally as a (1 x 200) array (dimension 1 is of size 1) and the first element of #BLOCKSIZE is 16, 15/16 of your GPU's processors will be idle while computing this layer. Similarly, if dimension 1 is of size 17, you will be wasting 15/32 of your processing power. This should be taken into consideration when assigning external dimensions to internal ones.
Note that dimensionality cannot be overridden by subtypes.
Mapping Dimensions to a Common Coordinate Space
The final dimension-related property, dmap, activates a powerful feature of the CNS framework. In networks where connectivity is assumed to be regular (i.e. cells do not have explicit lists of their inputs), a cell needs to be able to "find" its inputs, i.e., infer which cells in some other layer should be its inputs, based on its own position in its layer. This can be awkward for a number of reasons.
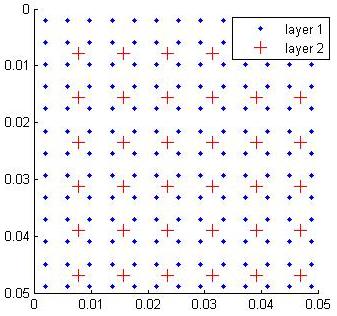
Consider this common case. Say we have two 2-D layers:
- Layer 1 is a 256x256 grid of pixels.
- Layer 2 is the result of a 4x4 convolution over layer 1, subsampled by a factor of 2. (The filter moves over layer 1 in steps of 2.) Due to the subsampling, and edge effects, layer 2 will be 127x127.
Even after one such operation, it is a little tricky to keep the proper cells in correspondence between the two layers. There are a number of cases where it gets worse:
- Working with an image pyramid having multiple scales, and trying to do pooling over scales.
- Trying to pool the results of two different pathways through a network, which have involved different steps, and now have different resolutions.
CNS's solution to this is to map dimensions to a common coordinate space that uses floating-point coordinates. A common choice is the interval [0, 1]. Consider the figure below for our two-layer example:
Now each cell in layers 1 and 2 is positioned in a common coordinate space. A layer 2 cell can find its inputs simply by asking for its four nearest neighbors in each dimension.
The dmap property controls which dimensions get mapped like this. Consider our 4-D example above. If we only have limited receptive fields over dimensions 'y' and 'x', we need only map those two dimensions:
p.dnames = {'f' 't' 'y' 'x'};
p.dims = {1 2 1 2};
p.dparts = {2 2 1 1};
p.dmap = [0 0 1 1];
When you are building a network model using mapped dimensions, the cns_mapdim function helps you assign the common coordinates.
Dimension Splitting
As explained above, CNS represents N-D structures (cell layers and array fields) internally as 2-D inside the GPU, often using GPU textures. However, current GPUs limit texture sizes to 65536 x 32768. If one of your dimensions is very large, this may cause problems.
For example, suppose we want to define a 3-D layer type which is going to have two spatial dimensions (y and x) and a feature dimension (f):
p.dnames = {'f' 'y' 'x'};
p.dims = {1 1 2};
p.dparts = {2 1 1};
Note that both f and y are assigned to internal dimension 1. If the maximum layer size along y is going to be 400, this would limit us to 163 features. The solution to this is a CNS feature called dimension splitting. We can use this alternate dimensionality definition:
p.dnames = {'f' 'y' 'x'};
p.dims = {[1 2] 1 2};
p.dparts = {[2 2] 1 1};
Now the f dimension is split across internal dimensions 1 and 2. This happens transparently; as a CNS programmer, you will still refer to it as a single dimension, both in MATLAB and in kernels. There are only two consequences to using a split dimension:
- There may be a slight performance hit.
- For CNS to represent the dimension internally, it must factor the dimension size you provide into two parts. The first part must (for implementation reasons) be a power of two. Thus, for split dimensions, it is best to choose dimension sizes that contain many factors of 2 so that the dimension can be split into two somewhat equal parts. For example, 5000 = 2354, so 5000 would be split into [8 625]. In our example, if the x dimension is also size 400, then internal dimension 2 would still have size 625 x 400 = 250,000 -- too large to fit into a GPU texture. A better choice would be, for example, 5120 = 21051, which would be factored into [64 80].
When you actually create a network model, you have two options:
- Easy: just give the total size of every dimension and let CNS decide how to factor any split dimensions. (The algorithm is in cns_splitdim.) You only have to ensure that the dimension size you use contains many factors of 2. For example:
m.layers{6}.size = {5120 400 400}; - Less easy: "manually" specify how dimensions are to be split. For example:
m.layers{6}.size = {[64 80] 400 400};
Fields
In a CNS model, a field is a numerical quantity that is associated with the model, a layer (or group of layers), a single cell, or a single synapse. All fields have an initial value at the time a model is initialized: either one provided by you at initialization or one defined as a default at package definition time. Some fields are variables that can change their value as a model runs inside the GPU; others can only be changed from MATLAB. All fields can be accessed from kernels using named macros.
Each field is defined by adding a line like this to the CNSFields method of the appropriate ".m" file.
d.fieldname = {class, modifiers};
For example:
d.V_m = {'cv', 'dflt', -70.0};
You choose the field's class from the following table. The class determines:
- The scope of the field: whether the field is associated with an entire model, a layer, a group of layers, a single cell, or a single synapse.
- Where you define it: most fields are defined in the appropriate package_type.m file, but fields with "model" scope are defined at the package level, i.e. the package.m file. (Note that subtypes inherit all the fields of their parent type.)
- Whether the field can be public (readable from any cell) or private (readable only from cells within the scope in which it is defined).
- The size of the field. Most fields can be defined to hold scalars or vectors. A few can hold entire N-D arrays; a common use for these is to store static feature dictionaries.
- Whether the field is variable (can be written to) while the model is running in the GPU or not.
In the above example, "'dflt', -70.0" is a modifier, supplying additional information about the field (in this case, a default value). The class determines which modifiers apply (see following sections).
| Class | Scope | Defined For | Public? | Size | Variable? | Description / How to Define |
| mp | model | package | yes | scalar or vector | no | Details. |
| mz | model | package | yes | scalar or vector | no | Details. |
| ma | model | package | yes | N-D | no | Details. |
| gp | group | type | define | scalar or vector | no | Details. |
| gz | group | type | no | scalar or vector | no | Details. |
| ga | group | type | define | N-D | no | Details. |
| lp | layer | type | define | scalar or vector | no | Details. |
| lz | layer | type | no | scalar or vector | no | Details. |
| la | layer | type | define | N-D | no | Details. |
| cc | cell | type | define | scalar or vector | no | Details. |
| cv | cell | type | define | scalar or vector | yes | Details. |
| sc | synapse | type | no | scalar or vector | no | Details. |
| sv | synapse | type | no | scalar or vector | yes | Details. |
Parameter Fields (classes: mp, gp, lp)
This class of fields is good for storing small parameters that apply to an entire layer (lp), group of layers (gp), or model as a whole (mp). They can only be set/reset from MATLAB; they cannot be changed as the model is running in the GPU. Internally they are stored in the GPU's constant cache, so kernels can access them quickly.
Required syntax:
|
Optional modifiers:
|
Examples:
This defines a parameter field that will be associated with a single layer. It will take a single integer value which, because there is no default, must be explicitly provided at initialization time. This defines a parameter field that will be associated with an entire model, takes multiple floating-point values, and defaults to the vector [0.1 0.2] if a value is not explictly provided at initialization time. |
Pointer Fields (classes: mz, gz, lz)
These are special parameter fields that hold the number of another layer in the model, i.e. that "point" to another layer. Kernels use these to read the values of fields in other layers.
Required syntax:
Where:
|
Optional modifiers:
|
Examples:
This defines a pointer field that will be set separately for each layer (of this type). It takes a single layer number and has no default value. The layer being pointed to can be of any type (because all types are subtypes of the 'base' type). This defines a pointer field that will be set separately for each layer (of this type). It can store a vector of layer numbers and defaults to []. Here, the layer(s) being pointed to must be of type 'input' (or a subtype). |
N-D Array Fields (classes: ma, ga, la)
N-D array fields allow you to share a large, N-dimensional array of data among all cells in a layer (la), group of layers (ga), or model as a whole (ma). A common usage is storing a large feature dictionary. These arrays can only be set/reset from MATLAB; they cannot be changed as the model is running in the GPU. Unlike parameter fields, they are stored in GPU main memory, which has high latency (but the 'cache' option can mitigate this).
Required syntax:
Where:
|
Optional modifiers:
|
Examples:
This defines a field that will be associated with a group of layers and will contain a 1-D array of integers. This defines a field that will be associated with a group of layers and will contain a 3-D array of floating-point numbers. Reads from the array will be cached using a GPU texture. |
Cell Fields (classes: cc, cv)
A cell field holds a scalar or vector value for each cell. Cell fields are stored in GPU main memory, which has high latency (but the 'cache' option can mitigate this).
- 'cv' fields ('cell variables') may change their values as the model runs inside the GPU. Each cell can only change its own value.
- 'cc' fields ('cell constants') are used to store per-cell constants. Their values can only be changed from MATLAB.
By default, cell fields are public -- a cell's value for the field can be read by other cells. Public 'cv' fields are typically used to store output values. To avoid race conditions, public 'cv' fields are double-buffered so that changes during one iteration of the network only become visible in the next iteration.
Cell fields can optionally be declared private, meaning they are not readable by other cells. Private 'cv' fields are useful for storing internal state variables in dynamic models, and private 'cc' fields can hold per-cell constants that are only used within each single cell.
Currently, only private cell fields can hold multiple (vector) values. The number of values must be the same for all cells in a layer.
Required syntax:
|
Optional modifiers:
|
Examples:
This defines a variable that will contain a floating-point number for each cell. Unless otherwise specified during initialization, it will start out with value -70.0 for each cell. The field is readable by other cells and cached for speed using a GPU texture. This defines a per-cell, multivalued, floating-point constant with no default value. Values for all cells will have to be provided at initialization time. Each cell can only read the value of its own constant. This defines a per-cell, scalar integer variable with a default value of 0. The field is not readable by other cells. |
Synapse Fields (classes: sc, sv)
Synapse fields are similar to private cell fields (above), except they are associated with each of a cell's explicit synapses. They can be used to keep track of separate constants and variables for each synapse.
Compile-Time Constants
A compile-time constant is a numerical quantity whose value is assigned at package definition time; it cannot be changed without modifying and recompiling the package. Compile-time constants can be defined in the same places as fields, and can be accessed both from MATLAB (using the cns_getconsts function) and from within kernels (by name).
A floating-point constant can be defined like this:
d.dt = 0.0001;
An integer constant is defined like this:
d.spk_dur = {11, 'int'};
Note: for every field with a default value, a compile-time constant is created called "fieldname_dflt". Conversely, defining such a constant is an alternative method for assigning a default value to a field.
Additional MATLAB Methods
Since a package directory already contains a set of ".m" files that form a type hierarchy (plus the package.m file), these files are a convenient place to put additional MATLAB methods.
Currently, the only such method that CNS will automatically call, if it is present, is a CNSInit method defined in the package.m file. It will be called just before a model is initialized on the GPU, and is a good place to put code that fills in some fields for you automatically. There is an example of this in the demo package; see the file cns/demopkg/demopkg.m. To call methods other than this one, you can use ordinary MATLAB method call syntax; however, in some cases it is safer to use the cns_call function.
Remember that CNS models run on the GPU, not inside MATLAB, so any additional MATLAB methods you create cannot affect a running CNS model (unless they call the cns function). They are, however, a good place to put code for setting up models.
To add a package-level MATLAB method, just add it to the package.m file. Type-specific MATLAB methods go in the appropriate package_type.m file. All such methods must be static.
A subtype can override a method provided by a supertype just by declaring a method of the same name. If the subtype's version needs to call the overridden version, it can do so using the function cns_super.
Writing Kernels
A kernel is a function that gets called separately for every cell in the network. Its job is to update the cell's variables; specifically, any cell variables (class cv) and synapse variables (class sv). In order to perform this computation, a cell's kernel can use any of the information it has access to: values of public fields in other cells, the current values of its own fields, parameters and N-D arrays defined at the model/group/layer level, etc.
Every type of cell has a single kernel, or no kernel. During a single network iteration, every cell (except those without a kernel) generally gets its compute kernel called once (although this can be changed). In some networks (e.g., spiking simulations) all the cells can be computed in parallel, while in other networks, layers must be computed in a specific order.
Kernels are written in C++, augmented by macros generated by CNS which mainly assist in reading and writing fields. Consider the following kernel, which is from the demo package. It applies a bank of different 2-D filters to an image, generating a 3-D result. Like all kernels, this code is responsible for computing only a single cell in the layer. CNS macros are shown in RED with names supplied by the package author in BOLD. The rest is ordinary C++.
#BLOCKSIZE 16 16 // Find coordinates of input cells in the previous layer. int y1, y2, x1, x2; FIND_LAYER_Y_NEAREST(PZ, FVALS_Y_SIZE, y1, y2); FIND_LAYER_X_NEAREST(PZ, FVALS_X_SIZE, x1, x2); // Iterate over input cells. float res = 0.0f; float len = 0.0f; for (int j = 0, x = x1; x <= x2; j++, x++) { for (int i = 0, y = y1; y <= y2; i++, y++) { // Read value of input cell. float v = READ_LAYER_VAL(PZ, 0, y, x); // Read corresponding filter value. float w = READ_FVALS(i, j, THIS_F); res += w * v; len += v * v; } } res = fabsf(res); if (len > 0.0f) res /= sqrtf(len); // Write out value of this cell. WRITE_VAL(res);
Kernel Files and Compilation
When cns_build compiles a package, it needs to compile a kernel for every non-abstract cell type. It starts by looking for the file package_type.h. If that file doesn't exist, cns_build will look for the supertype's .h file, and so on up the hierarchy.
It is also possible for an abstract cell type to define a "template" kernel which contains placeholders that get filled in with code provided by a subtype. See the #PART and #INCLUDEPART preprocessor directives.
C++ Usage in Kernels
When you write a kernel, you are writing the body of a function; your code will actually be embedded inside a function before it is compiled. Hence, you can only write C++ that is valid inside a function body. (If you need to write any auxiliary helper functions, click here.)
Most standard C++ mathematical functions are supported.
It is important to note that GPUs are only fast for 32-bit quantities (or smaller). Your code should mainly use the datatypes float and int. Avoid the double type.
- Use the single-precision version of floating point functions, e.g., sqrtf(x) instead of sqrt(x).
- Floating-point constants should end in "f", for example, 0.0f.
For details on the speed of various mathematical operations on the GPU, refer to the CUDA Programming Guide. One operation that is particularly slow is integer division / modulo.
CNS Preprocessor Directives
The following two preprocessor directives are used frequently. Click here for others.
#BLOCKSIZE
This directive must appear at the beginning of your kernel. It defines the block size, which is (more or less) how many cells in a layer the GPU will attempt to compute simultaneously. The maximum block size depends on (a) the complexity of your kernel and (b) the resources on your GPU. You will generally want to make the block size as large as possible; however, setting it too high will cause an error, so some experimentation is required. A good first guess is usually (16 x 16). The first element must be a multiple of 16, so usually you will be adjusting the second. The syntax is:
For example:
#NULL
Sometimes you may need to define a cell type that doesn't do any computation. This only makes sense for cells that are going to represent inputs to a model. You can do this by writing a kernel consisting of the single line:
CNS Macros
This section describes all the macros that CNS generates for you to use in a kernel of a given type. Most of these are based on the fields accessible from that kernel, which include:
- Fields defined in this type.
- Fields defined in any supertypes.
- Fields defined at the package level, i.e. having "model" scope.
- Fields defined in other types to which we have pointer fields or explicit synapses.
Macro names are always entirely UPPER case, regardless of the case of your definitions. This makes it easier to distinguish CNS macros from regular C++.
Note: the cns_build function has a help option that lists all the macros available to each cell type's kernel.
General
| Macro(s) | Description |
ITER_NO |
The current network iteration number. Initialized to 1 by the cns('init') command. Automatically incremented by the cns('run') command. Can also be queried or changed from MATLAB using the cns('get') and cns('set') commands. (Note: ITER_NO is zero-based in kernels, but one-based in MATLAB.) |
PRINT(format, ...) |
Use this instead of C++'s printf function to print output to the terminal. Note that this only works when running CNS in debug mode. Uses the same syntax as printf. |
ERROR(format, ...) |
Just like PRINT above, except that it halts execution. As with PRINT, it only works in debug mode. |
CNS_INTMIN CNS_INTMAX CNS_FLTMIN CNS_FLTMAX |
The minimum and maximum values of the int and float datatypes in CNS, respectively. CNS also provides equivalent MATLAB functions (same names but lower case) that return these values. |
Connectivity: Regular Grid
The following macros are used in kernels that assume regular grid connectivity among cells.
| Macro(s) | Description |
type_PTR |
This is a C++ class that can hold a pointer to a layer of a particular type of cells. Use this class if you need to define a local (C++) variable to hold such a pointer (whose value would be retrieved using one of the macros immediately below). Such pointers are used as inputs to many other macros.
For example (assuming a cell type called "weight" and a multivalued pointer field called "pzs" that contains pointers to "weight" layers): Of course, often you might not want to define a local variable. You could also do this: int yc = WEIGHT_Y_SIZE(PZS(i)); |
THIS_Z |
Returns a type_PTR to the current layer, where type is the current type. |
field field(e) |
Returns a type_PTR to another layer, where type is the type of that layer.
|
NUM_field |
The number of values in a multivalued pointer field.
|
dim_SIZE type_dim_SIZE(z) |
Return the number of cells along a particular dimension of this or another layer.
|
THIS_dim |
Zero-based integer coordinate of this cell in its layer along a particular dimension.
|
FIND_type_dim_NEAREST(z, n, &v1, &v2) FIND_type_dim_WITHIN (z, r, &v1, &v2) FIND_type_dim_NEAREST(z, n, &v1, &v2, &i1, &i2) FIND_type_dim_WITHIN (z, r, &v1, &v2, &i1, &i2) |
Find a range of (zero-based integer) coordinates of other cells that are close to this cell in common coordinate space along a particular dimension.
The first two macros return false if the returned range (v1 - v2) is smaller than expected, i.e., had to be adjusted so as not to contain invalid coordinates. The last two macros return false if the returned range (v1 - v2) is empty, i.e., no valid coordinates were found. Note: cns_findnearest and cns_findwithin are the equivalent MATLAB functions. |
THIS_dim_CENTER type_dim_CENTER(z, c) |
Return the position in common coordinate space of this cell or a cell in another layer along a particular dimension. This is a floating point number.
Note: cns_center is the equivalent MATLAB function. |
FIND_type_dim_NEAREST_AT(z, p, n, &v1, &v2) FIND_type_dim_WITHIN_AT (z, p, r, &v1, &v2) FIND_type_dim_NEAREST_AT(z, p, n, &v1, &v2, &i1, &i2) FIND_type_dim_WITHIN_AT (z, p, r, &v1, &v2, &i1, &i2) |
These macros are the same as the above FIND_type_dim_... macros, except they find cells near a specified point in common coordinate space (along the relevant dimension). They take one additional parameter:
Note: cns_findnearest_at and cns_findwithin_at are the equivalent MATLAB functions. |
Connectivity: Explicit Synapses
The following macros are used in packages where cells are connected via explicitly-enumerated synapses.
| Macro(s) | Description |
NUM_SYN |
The number of explicit synapses this cell has. An integer. |
SELECT_SYN(e) |
Makes a particular synapse "active", which means that all macros that reference fields of presynaptic cells or synapse fields will refer to this synapse (until the next SELECT_SYN call).
|
SYN_Z |
Returns a pointer to the layer in which the currently active presynaptic cell (as determined by SELECT_SYN) resides. Not required if you only want to read fields of presynaptic cells or synapse fields, but useful if you want to read other information from the presynaptic layer (using macros in this section). |
SYN_dim |
Returns the zero-based integer coordinate (along a particular dimension) of the currently active presynaptic cell in its layer. Not required if you only want to read fields of presynaptic cells or synapse fields, but useful if you want to read information from other cells in the presynaptic layer (using macros in this section).
|
Parameter Fields
| Macro(s) | Description |
field field(e) type_field(z) type_field(z, e) |
Return the value of a parameter field defined for (a) the model as a whole, (b) this layer, or (c) another layer.
|
NUM_field NUM_type_field(z) |
The number of values in a multivalued parameter field.
|
Cell Fields
| Macro(s) | Description |
READ_field READ_field(e) |
Return the value of a cell field (for this cell).
|
WRITE_field(v) WRITE_field(e, v) |
Update the value of a cell variable (for this cell).
|
NUM_field |
The number of values in a multivalued cell field. Note that this number will be the same for all the cells in a layer.
|
READ_type_field(z, c1, c2, ...) READ_PRE_field |
Return the value of a public cell field for an arbitrary cell, or for the currently selected presynaptic cell.
Note: the READ_type_field macro is often used inside loops which can benefit from optimization. See this section for faster alternatives. |
Synapse Fields
| Macro(s) | Description |
SYN_TYPE |
Return the synapse type of the currently selected synapse.
Note that synapse types are 0-based in kernels but 1-based in MATLAB. |
READ_field READ_field(e) |
Return the value of a synapse field for the currently selected synapse.
|
WRITE_field(v) WRITE_field(e, v) |
Update the value of a synapse variable for the currently selected synapse.
|
NUM_field |
The number of values in a multivalued synapse field. Note that this number will be the same for all synapses of all the cells in a layer.
|
N-D Array Fields
| Macro(s) | Description |
field_dim_SIZE field_dim_SIZE(e) type_field_dim_SIZE(z) type_field_dim_SIZE(z, e) |
Return the number of cells along a particular dimension of an N-D array field belonging to (a) the model as a whole, (b) this layer, or (c) another layer.
|
READ_field(c1, c2, ...) READ_field(e, c1, c2, ...) READ_type_field(z, c1, c2, ...) READ_type_field(z, e, c1, c2, ...) |
Return a value from an N-D array field.
Note: these macros are often used inside loops which can benefit from optimization. See this section for faster alternatives. |
NUM_field NUM_type_field(z) |
Return the number of N-D arrays held in a multivalued N-D array field.
|
Compile-Time Constants
CNS also creates macros for every compile-time constant a kernel has access to: those defined at the package level, in a supertype, or for the type itself.
The macro names are just UPPER case versions of the constant names. For example, a compile-time constant called "spk_dur" will be accessible using macro SPK_DUR.
Optimization: Public Cell Fields
It is common to have kernels that read the values of many cells of the same layer inside a loop, using the READ_type_field macro. CNS provides two somewhat faster sets of macros for doing this.
The first set of macros introduces the concept of a handle. In your code, instead of this:
for (...) {
...
float v = READ_type_field(z, c1, c2, ...);
...
}
you would do this:
[type_]field_HANDLE h = GET_type_field_HANDLE(z);
...
for (...) {
...
float v = READ_[type_]field_HANDLE(h, c1, c2, ...);
...
}
Internally, the handle points to the block of memory that contains that field of layer z (for all cells). This saves having to find that address for each loop iteration. Once you have the handle, you can use handle-based macros in place of macros that take z.
| Macro(s) | Description |
[type_]field_HANDLE |
This is a C++ class that holds a handle (described above) to a public cell field in a layer. You get a handle using this macro.
|
GET_type_field_HANDLE(z) |
Returns a handle to a public cell field in a particular layer. The handle can then be passed as a parameter to the macros below.
|
READ_[type_]field_HANDLE(h, c1, c2, ...) |
Returns the value of a public cell field for a cell in a layer, using a handle.
|
[type_]field_HANDLE_dim_SIZE(h) |
Returns the number of cells along a particular dimension of the layer pointed to by a handle.
|
The second set of macros makes a further optimization, but requires understanding how an N-D layer of cells is mapped internally to 2-D. Normally when reading the value of a cell's field, you have to provide a full set of N-D coordinates (c1, c2, ...) to identify the cell; CNS automatically converts these to 2-D coordinates and then performs the lookup. The following macros let you work directly in the internal 2-D space.
| Macro(s) | Description |
GET_type_field_IPOS(z, c1, c2, ..., &y, &x) |
TODO |
GET_[type_]field_HANDLE_IPOS(h, c1, c2, ..., &y, &x) |
TODO |
READ_[type_]field_IPOS(y, x) |
TODO |
Optimization: N-D Array Fields
The following optimization macros for reading from N-D array fields are very similar in concept and syntax to those for reading the fields of cells in other layers, described above. See that section for an explanation of the concepts.
| Macro(s) | Description |
[type_]field_HANDLE |
This is a C++ class that holds a handle to an N-D array field. You get a handle using one of these macros.
|
GET_field_HANDLE GET_field_HANDLE(e) GET_type_field_HANDLE(z) GET_type_field_HANDLE(z, e) |
Return a handle to an N-D array field belonging to (a) the model as a whole, (b) this layer, or (c) another layer. The handle can then be passed as a parameter to the macros below.
|
READ_[type_]field_HANDLE(h, c1, c2, ...) |
Returns a value from an N-D array field, using a handle.
|
[type_]field_HANDLE_dim_SIZE(h) |
Returns the size (along a particular dimension) of the N-D array field pointed to by a handle.
|
| Macro(s) | Description |
GET_field_IPOS(c1, c2, ..., &y, &x) GET_field_IPOS(e, c1, c2, ..., &y, &x) GET_type_field_IPOS(z, c1, c2, ..., &y, &x) GET_type_field_IPOS(z, e, c1, c2, ..., &y, &x) |
For cached N-D array fields only. TODO |
GET_[type_]field_HANDLE_IPOS(h, c1, c2, ..., &y, &x) |
TODO |
READ_[type_]field_IPOS(y, x) |
For cached N-D array fields only. TODO |
READ_[type_]field_IPOS(h, y, x) |
For uncached N-D array fields only. TODO |
More CNS Preprocessor Directives
#PART and #INCLUDEPART
These directives make it easy to create a number of similar kernels without duplicating code. Consider the following example, in which we want to create two kernels, both of which run 2-D filters over a layer. The only difference is in the particular mathematical function that is computed between a filter and the patch of the layer it overlays. Using #PART and #INCLUDEPART, we specify the general form of the algorithm once, in an abstract supertype, with placeholders that get filled in by the subtypes.
Example (abstract) supertype kernel. #INCLUDEPART lines are placeholders which will be replaced by code from subtypes:
#BLOCKSIZE 16 16 int y1, y2, x1, x2; FIND_LAYER_Y_NEAREST(PZ, FVALS_Y_SIZE, y1, y2); FIND_LAYER_X_NEAREST(PZ, FVALS_X_SIZE, x1, x2); float res = 0.0f; for (int j = 0, x = x1; x <= x2; j++, x++) { for (int i = 0, y = y1; y <= y2; i++, y++) { float v = READ_LAYER_VAL(PZ, 0, y, x); float w = READ_FVALS(i, j, THIS_F); #INCLUDEPART update } } #INCLUDEPART done WRITE_VAL(res);
Example subtype kernel #1 (convolution). Here, #PART is used to identify code used to fill in each placeholder.
#PART update res += w * v; #PART done // Nothing.
Example subtype kernel #2 (distance):
#PART update float diff = w - v; res += diff * diff; #PART done res = sqrtf(res);
Subtype kernels defined using #PART can also override the parent type's #BLOCKSIZE.
These directives can be used recursively: a #PART section can itself contain #INCLUDEPART lines which will insert code from a yet more specific subtype.
Loop Unrolling
CNS provides a smart loop unrolling mechanism that is useful for speeding up tight inner loops. It is "smart" in that the unrolling will not affect program correctness if, at runtime, you need a number of iterations that is not an integer multiple of the unroll factor.
To use loop unrolling, replace your C++ for statement with one of these #UNROLL_START directives:
This is semantically equivalent to the following for line: for (symbol = lower; symbol cond upper; symbol++) {factor
The unroll factor, i.e. the number of iterations that the preprocessor will group into a single iteration.
symbol
A symbol within the body of the loop that will be replaced by the value of the loop counter. Make sure it is a distinctive string of characters that isn't a substring of other identifiers inside the loop. For example, use %i% rather than just i.
Note: if none of the following parameters are given, the loop will always execute exactly factor times.
lower
The starting value of the loop counter. Can be a constant or the name of a local C++ variable. Defaults to 0.
cond
Either < or <=. Defaults to <.
upper
The upper bound of the loop counter. Can be a constant or the name of a local C++ variable.
You must also replace the closing } of the for loop with the directive #UNROLL_END, and replace any break statements with #UNROLL_BREAK. Both these directives must appear on their own lines.
Fast Local Array Variables
When you declare a local (C++) variable in a kernel, the CUDA compiler usually assigns it to a register, which is fast. However, local variables that are arrays usually get assigned to global GPU memory, which is much slower. CNS provides a way to declare small local fixed-size arrays that get stored in shared memory, which is as fast as registers. (Note that while they reside in "shared" memory, this is only for speed reasons. They have the same scope as other local variables, i.e., they are temporary variables that exist only for the duration of a single cell's kernel call.)
Fast local arrays are declared at the beginning of your kernel.
Here are some example local array definitions:
#ARRAY gcond 10 (a 10-element array of floats) #ARRAY syn_t 5 int (a 5-element array of ints) #ARRAY xyz 8 double (an 8-element array of doubles)
Within kernels you access array elements using macro syntax. For example:
GCOND(3) = 3.6f; float gc = GCOND(0); for (int i = 0; i < 5; i++) SYN_T(i) = 0; ...
Note that the size of shared memory is limited (to 16KB in current GPUs), and using a lot of arrays will mean you have to make the cell type's block size smaller.
Helper Functions
A cell type's compute kernel is the body of a single function. If you want to have a library of auxiliary "helper" functions that can be called from different kernels, you can put these in the file package.h. in your package directory.
Here is an example package.h file that contains two helper functions:
INLINE float Sigmoid(float x) { return 1.0f / (1.0f + expf(-x)); } INLINE float DSigmoid(float y) { return y * (1.0f - y); }
Each function definition must start with the word INLINE.
Checking Kernel Memory Usage
Once you have written a kernel, you may want to check to see how efficiently it compiles for the GPU. The cns_build function has an info option that displays some useful information, such as:
- Number of registers used (less is better).
- Number of local variables that would not fit into registers (preferably none).
See the cns_build function for details.